Humanness Prediction¶
Humanness prediction is the task of predicting how much an antibody sequence looks like a human antibody sequence.
This model has been integrated into the "Antibody Optimization" job
When you create an Antibody Optimization job, the humanness scores and germline sequence identities of the input antibodies will be automatically calculated.
Features¶
- Accurate metric: Efficient and accurate humanness metric based on big data of human antibody sequences in OAS (Observed Antibody Space).
- Intuitive interface: Major results are clear and easy to read. Compare with germline sequences and beware of the risk sites on one page.
- Cloud-based, mass-scale: Support batched prediction of multiple sequences on our cloud platform. Enjoy high accuracy and high efficiency at the same time!
Inputs¶
To submit an Humanness Prediction job, open the Project Editor and click "New Job" button on the left sidebar. Then click "Humanness Prediction" under the "Antibody Design" group to open the job submission page.
- Antibodies:A list of antibody sequences in FASTA format. You can either enter the sequences in the input box or upload a FASTA file (by clicking the "
" button). Each antibody is predicted independently.
- Chain type: Chain type(s) of the input antibodies. Could be either "VH", "VL" or "VH+VL" (default). If you want to predict humanness for nanobodies, please select "VH" here.
- Antibody Name: Name of the antibody. Defaults to "Antibody". To change it, hover above the antibody name, and click on the "
" button.
- Sequence: Sequence of an antibody. Each chain shall not exceed 200 residues.
(Upload Sequence): Upload your own antibodies (a .FASTA file) by clicking the "
" button. Each antibody must have 2 chains (if the chain type is "VH+VL") with labels sharing the same prefix and ending with "_H" and "_L" respectively. Above is an example.
- Job Name: Name of the job. Note that the job name must be unique within the project.

Model & Parameters¶
GeoHumAb, our proprietary humanness prediction model, is available for this job. Parameters of this model are as follows.
- scheme: Antibody numbering scheme. Could be either "Kabat", "IMGT", "Chothia" or "AHo".
- CDR def.: Numbering scheme used for definition of CDR regions. Could be either "Kabat", "IMGT", "Chothia", "North".
Results¶
Click the job name in the Files & Jobs panel to view the job results.
Result Summary¶
The result summary is stored in a CSV file, which can be downloaded by clicking the "" button to the top-right of the Summary Table.
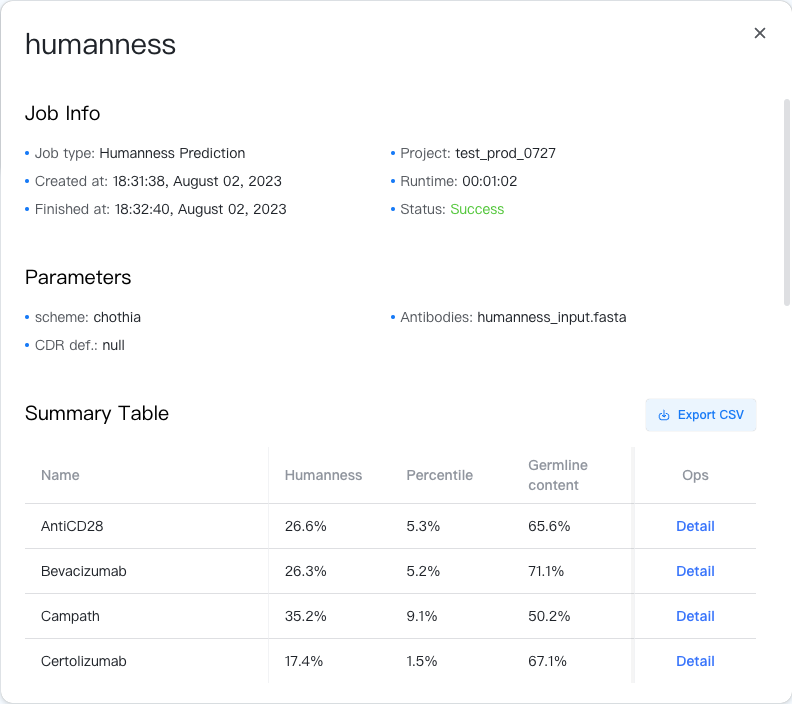
The summary table contains the following columns:
- Name: Name of the antibody.
- Humanness: Humanness score of the antibody, determined by the average occurrence of the 9-mer peptide fragments among OAS human assays. The higher the score, the more human-like the antibody is.
- Percentile: Percentile of the humanness score among all antibodies in OAS. The higher the percentile, the more human-like the antibody is.
- Germline content: Germline content of the antibody, determined by the sequence identity with nearest heavy and light human germline sequences.
In the rightmost column, you could click on the "Detail" button to view the detailed humanness results of the antibody.
Result Detail¶
The result detail page is divided into 3 sections: Summary, Germline Comparison and Detailed View. If the input chain type is "VH+VL", the Germline Comparison and Detailed View sections will be different for the heavy and the light chain. You can switch between the two chains by using the "" chain switcher just below the Summary section.
Raw data of the result detail is stored in a CSV file, which can be downloaded by clicking the "" button to the top-right of the Detailed View section.
Summary¶
The Summary section presents the model and the scheme used to calculate humanness. It also shows the humanness score, percentile, and germline content of the whole antibody, calculated by an average of the heavy and light chain values, if applicable.
Sequence Viewer¶
The Sequence Viewer sections aligns the target antibody chain with its closest human germline sequences. The name of these germline sequences are shown on the right. Germline residues that are identical to the target sequence are highlighted in blue.
Target residues with a germline frequency lower than 1% are marked with a red triangle.
Target residues that lie in a high-risk peptide fragment are highlighted in red. The more high-risk peptides a residue is in, the darker its color.
CDR residues are underlined with dark gray lines. Due to VD(J) recombination and somatic hypermutation, CDR regions are highly variable and thus are usually highlighted in red. However, this does not necessarily mean that these regions are immunogenic.
Vernier residues are underlined with light gray lines.
Terms
- Germline frequency: The residue frequency in the germline family at given position in human repertoires from the OAS database. The higher the frequency, the more likely the residue is a valid residue in the germline family.
- High-risk peptide: A 9-mer peptide is considered high-risk if less than 10% of human assays in OAS contain this peptide.
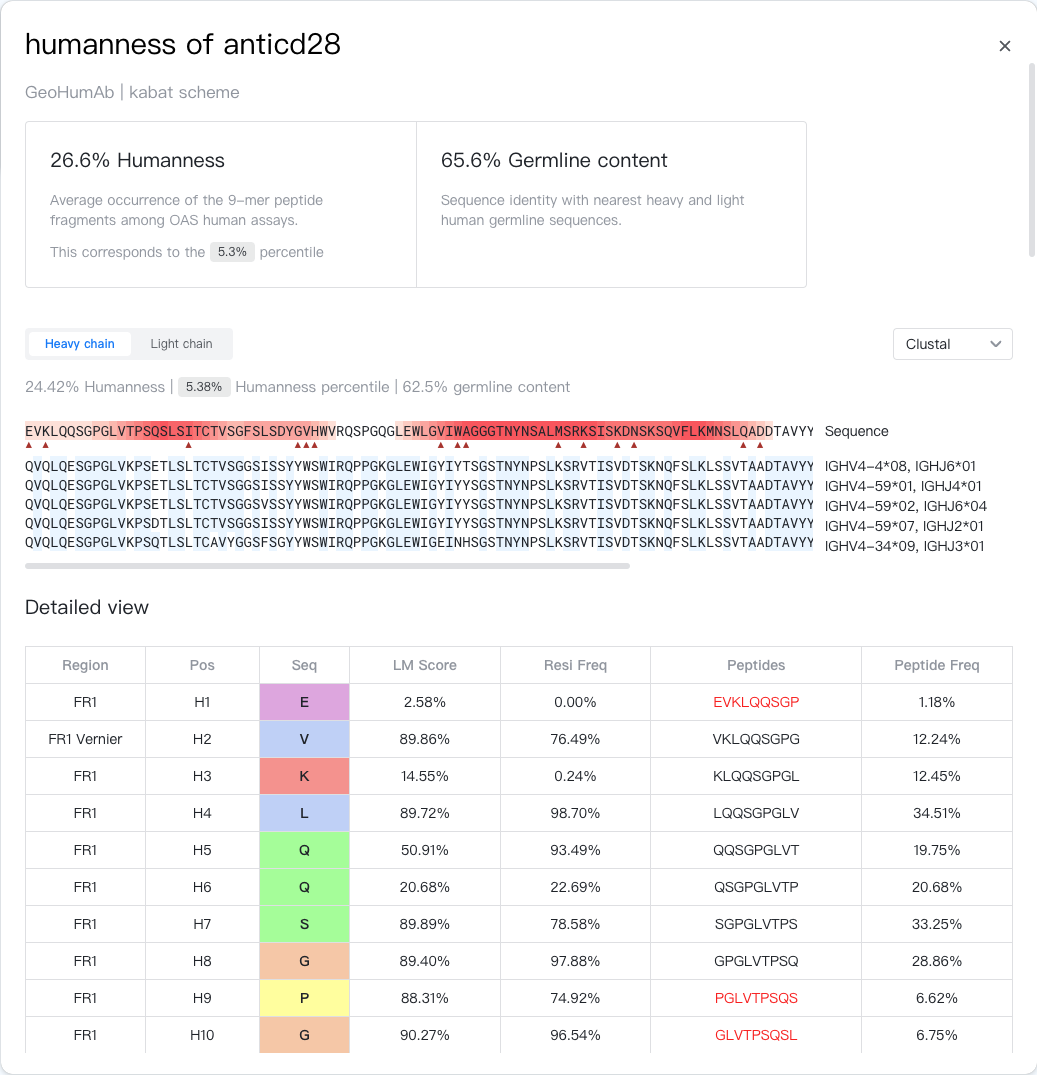
Detailed View¶
The Detailed View section presents the detailed metrics for each position in the antibody chain in a table. The table contains the following columns:
- Region: Antibody region this position belongs to according to the CDR definition. Could be FR1-4 / CDR1-3. Vernier and VH-VL interface regions are also shown.
- Pos: Position in the antibody chain, numbered by the specified scheme. Shown in the format of "{chain type}{residue number}{insertion code}", e.g. "H100A".
- Seq: Residue type at this position. Residues are colored by a speicified color scheme. You can change the scheme using the "
" color scheme selector at the top-right of the table.
- LM Score: Language model score of the residue. The higher the score, the more likely the model thinks this residue is the most suitable one in this context.
- Resi Freq: Residue frequencyin the germline family at given position in human repertoires from the OAS database. The higher the frequency, the more likely the residue is a valid residue in the germline family.
- Peptides: The 9-mer peptide that starts with this residue. If the corresponding "Peptide Freq" is below 10%, the peptide is shown in red and is considered a high-risk peptide.
- Peptide Freq: The frequency of the peptide occuring in a OAS human assay.