Affinity Prediction¶
Affinity prediction is the task of predicting the binding affinity of an antibody-antigen complex in terms of \(K_D\) (mol).
Challenges¶
- Noisy data: Public affinity data comes from multiple laboratories with various experimental conditions, making them often incomparable. This poses an obstacle to training high-quality models.
- Inaccurate model: Physical simulation based models struggle to balance accuracy and compute cost. Many machine learning studies partition the data in a trivial way, resulting in models with inflated results that do not meet the needs of real-world use cases.
Features¶
- Precise prediction: Our model is pre-trained on large-scale protein structural data, modeling the input at both the atomic and residue level, integrating geometric information such as angles and dihedral angles, thus can accurately predict the affinity between antigens and antibodies.
- Cloud-based, mass scale: Supports batched prediction of multiple structures on our cloud platform. Enjoy high accuracy and high efficiency at the same time!
Inputs¶
To submit an Affinity Prediction job, open the Project Editor and select "Affinity Prediction" from the "Characterization" dropdown menu.
- Complex: Target antibody-antigen complex in PDB/mmCIF format. The structure can be uploaded from a local machine, imported from a cloud database or generated by a GeoBiologics job.
- Job Name: Name of the job. Note that the job name must be unique within the project.

Models & Parameters¶
GeoAffinity, our proprietary affinity prediction model, is available for this job. No additional parameters need to be provided.
Results¶
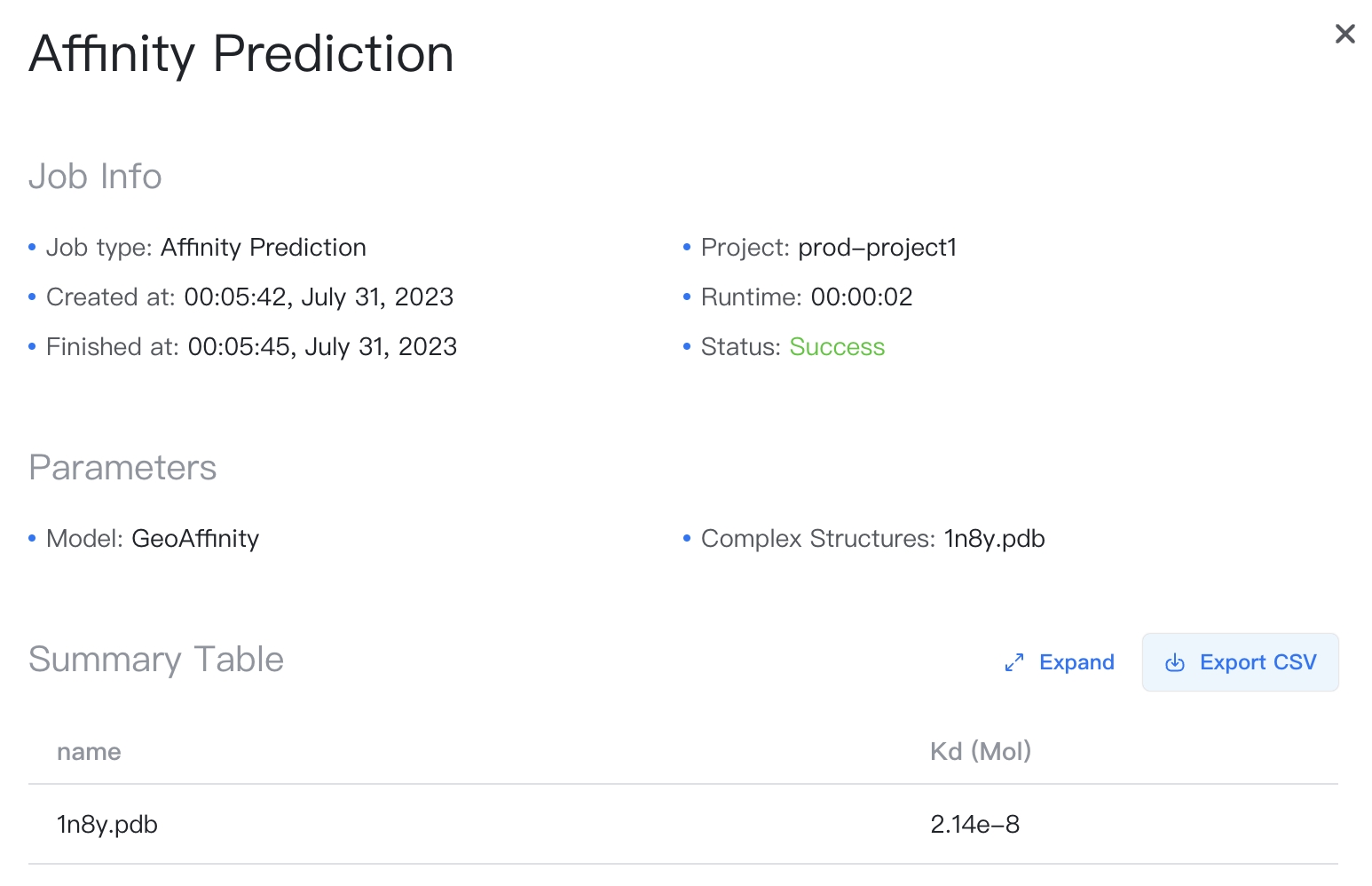
Click the job name in the Files & Jobs panel to view the job results.
The result summary is stored in a CSV file, which can be downloaded by clicking the "" button to the top-right of the Summary Table.
The summary table contains the following columns:
- name: Name of the complex, as in input.
- \(K_D\) (mol): Predicted binding affinity for the complex.