Antibody Optimization¶
Evaluate and optimize your lead antibodies with powerful and specialized AI assistants, namely
- Scorer: an AI that scores each amino acid in the antibody based on various properties, such as humanness, acidity and hydrophobicity.
- Mutator: an AI that mutates the input antibody sequence to form the initial output.
- Advisor: an AI that guides the optimization process by suggesting mutations that are likely to improve the antibody.
- Structure Modeler: an AI that predicts the structure of the antibody to enable interpretable, structure-based analysis.
The AI assistants are powered by the following models:
- BioGeometry's proprietary antibody structure prediction model,
- BioGeometry's proprietary antibody humanization and humanness prediction model,
- Evolutionary-scale protein language model (ESM) by Meta AI for protein sequence modeling and optimization,
- Side chain pKa and hydrophobicity prediction model,
- Sequence liability analysis model that predicts post-translational modifications (PTMs), along with other sequence liabilities.
Inputs¶
To submit an Antibody Optimization job, open the Project Editor and select "Antibody Optimization" from the "Optimization" dropdown menu.
- Antibody: The input lead antibody. You can either enter the sequences in the input box or upload a FASTA file (by clicking the "
" button).
- Antibody Name: Name of the antibody. Defaults to "Antibody". To change it, hover above the antibody name, and click on the "
" button.
- Sequence: Sequence of an antibody. Each chain shall not exceed 200 residues.
(Upload Sequence): Upload your own antibody (a .FASTA file) by clicking the "
" button. Your antibody must have 2 chains with labels sharing the same prefix and ending with "_H" and "_L" respectively. Above is an example.
- Antibody Name: Name of the antibody. Defaults to "Antibody". To change it, hover above the antibody name, and click on the "
- Job Name: Name of the job. Note that the job name must be unique within the project.

Models & Parameters¶
Below is a breakdown of the available models and parameters in Antibody Optimization.
- General parameters
- Scheme: Antibody numbering scheme. Could be either "Kabat", "IMGT", "Chothia" or "AHo".
- CDR def.: Numbering scheme used for definition of CDR regions. Could be either "Kabat", "IMGT", "Chothia", "North". Defaults to the same as scheme. Required when
scheme = AHo.
- Scoring parameters
- Scorer: AI assistant that scores each amino acid in the antibody based on various properties, such as humanness, acidity and hydrophobicity. Available models include:
- GeoHumanness: State-of-the-art humanness prediction model.
- Side-chain pKa: structure-based side-chain pKa prediction model. When using this model, the "Relax structure" parameter will be automatically set to "Yes".
- wwHydrophobicity (dG water-membrane): Wimley–White whole residue hydrophobicity scale that measures the free energy of transfer of unfolded chains from water to the bilayer interface
- wwHydrophobicity (dG water-octanol): Wimley–White whole residue hydrophobicity scale that measures the free energy of transfer of unfolded chains into octanol, which is relevant to the hydrocarbon core of a bilayer.
- wwHydrophobicity (dG difference): Difference between the dG water-membrane and dG water-octanol scores.
- Advisor: AI assistant that guides the optimization process by suggesting mutations that are likely to improve the antibody. Available models include:
- ESM1v-650M: Language model specialized for prediction of variant effects. Enables SOTA zero-shot prediction of the functional effects of sequence variations.
- ESM2-3B: SOTA general-purpose protein language model.
- GeoHumAb: Our proprietary, state-of-the-art humanization algorithm.
- Scorer: AI assistant that scores each amino acid in the antibody based on various properties, such as humanness, acidity and hydrophobicity. Available models include:
- Mutation parameters
- Mutator: AI assistant that mutates the input antibody sequence to form the initial output. Available models are shown below. Refer to the Humanization page for details on parameters of these models.
- None: No automatic mutation. The input antibody is used as the initial output.
- GeoHumAb: Our proprietary, state-of-the-art humanization algorithm.
- CDR Grafting: Traditional humanization technique that grafts the CDR regions of the input antibody onto human germline sequences.
- Mutator: AI assistant that mutates the input antibody sequence to form the initial output. Available models are shown below. Refer to the Humanization page for details on parameters of these models.
- Structure prediction parameters
- Relax structure: Whether to relax the model-generated antibody structure using Amber. Default to false. Must be true if using the side-chain pKa scorer.
Result Panel¶
Click the job name in the Files & Jobs panel to open the job result panel. This is where you perform interactive antibody optimization with the help of the AI assistants.
The input antibody sequence is shown in the line labeled "Parental (wt)". If a mutator is specified, the initial output will be generated by the mutator. Otherwise, it will be the same as the input antibody sequence. The current output antibody sequence is shown in the line labeled "Current (mt)".
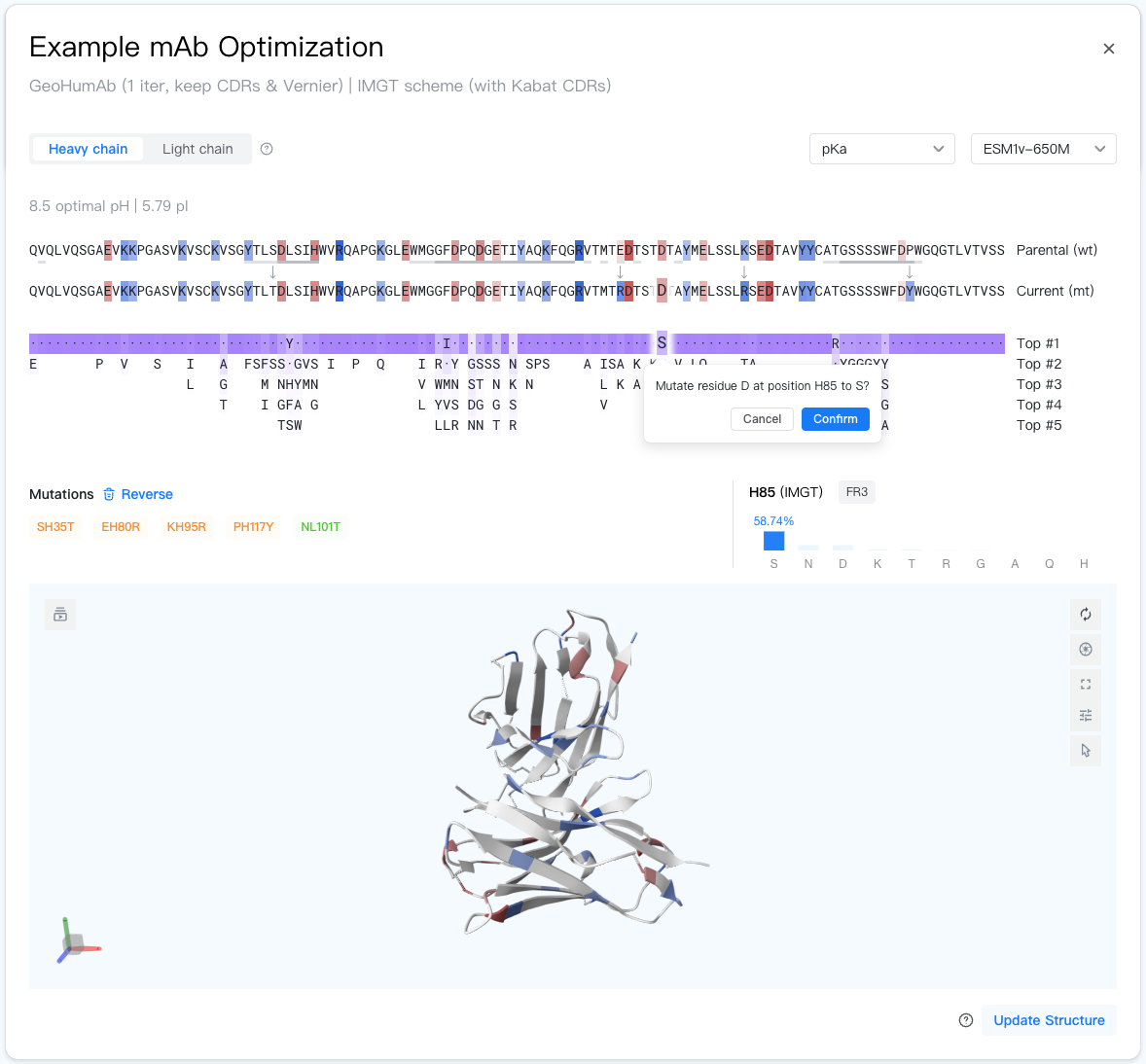
Job Summary¶
The job name and summary is shown at the top of the result panel as the title and subtitle. The job summary displays the mutator model and scheme/CDR definition used in this job. These parameters can not be changed after the job is submitted.
Sequence Viewer¶
Right below the job summary is the Sequence Viewer, which displays the parental and current antibody sequences and allows for interactive optimization.
At the top-left, you can switch between the two antibody chains by using the chain switcher "".
At the top-right, you can change the model for the AI scorer and advisor via the selectors.
See the Models & Parameters section for details on these models.
Visualizing the per-residue score¶
In the middle lies the parental and current antibody sequences, with the background color indicating the score of each amino acid residue. Positions where the parental and current sequences differ are marked with an arrow (↓).
If the scorer is GeoHumanness, current residues with a germline frequency lower than 1% are marked with a red triangle. If a parental residue has a germline frequency lower than 1% but its current counterpart has a germline frequency higher than 1%, it is marked with a green triangle.
CDR residues are underlined with dark gray lines. Vernier residues are underlined with light gray lines if using the Kabat CDR definition.
What is the coloring scheme for each scorer?
-
GeoHumanness: white-red.
Darker red indicates higher immunogenicity risk.
More concretely, a 9-mer peptide is considered high-risk if less than 10% of human assays in OAS contain this peptide. The more high-risk peptides a residue is in, the darker its color.
-
Side-chain pKa: blue-white-red.
Darker red indicates lower pKa (more acidic). Darker blue indicates higher pKa (less acidic).
-
wwHydrophobicity: cyan-white-yellow.
Darker yellow indicates higher hydrophobicity. Darker cyan indicates lower hydrophobicity.
Applying mutations with the help of AI advisor¶
Below the parental and current antibody sequences are the advisor suggestions. For each position, the advisor displays the top 5 likely amino acid residues (the current residue is also included in the ranking), with darker purple background indicating higher confidence.
To make the suggestions more readable, residues in the current sequence are shown as dots ("·") and mutations with less than 1% confidence are omitted.
When you hover on an advisor suggestion, the detailed information of the suggestion will be shown at the bottom right of the Sequence Viewer. It inclues:
- Position: The position you are hovering on, numbered by the specified scheme. Shown in the format of "{chain type}{residue number}{insertion code}", e.g. "H100A".
- Region: Antibody region this position belongs to according to the CDR definition. Could be FR1-4 / CDR1-3. If the CDR definition is set to Kabat, Vernier regions are also shown.
- Advisor confidence: The confidence of top 10 advisor suggestions, with the target residue type highlighted in blue. The higher the confidence, the more likely the mutation is to improve the antibody.
To apply an advisor-suggested mutation, simply click on the mutation suggestion and then click on the "Confirm" button in the popup. You can also initiate an arbitrary mutation by clicking on the residue you want to mutate in the "Current (mt)" sequence and then select a mutation from the popup.
Managing mutations¶
The mutations you have applied are shown in the "Mutations" section. Click on the "Reverse" button to select mutations to reverse.
Structure Viewer¶
The Structure Viewer displays the predicted structure of the current antibody. While this result panel is open, the Structure Objects panel at the bottom left of the webpage will be automatically synced to this panel.
Residues in the structure are colored by the same scheme as the Sequence Viewer, according to the AI scorer.
Since the inference cost for structure prediction is relatively high, the structure is only updated when you click on the "Update Structure" button at the bottom right, or when you apply a mutation while the scorer is set to "Side-chain pKa". Note that the residue coloring is always synced with the current (latest) antibody sequence, even if the structure corresponds to an older sequence.