Enzyme Mining¶
The Enzyme Mining aims to identify high-quality starting enzymes that can efficiently catalyze specific reactions from a rich library of enzymes. GeoBiologics focuses on key enzyme characteristics, including Enzyme Activity, pH Stability, and Thermostability and Stereoselectivity and Solubility, to ensure the selected enzymes perform excellently in practical applications.
Challenges & Features¶
The challenges faced during the enzyme mining process lie in accurately identifying enzymes suitable for specific reactions from a vast library, while considering multiple performance indicators. To address this, GeoBiologics’ enzyme mining module offers the following advantages:
-
Advanced Algorithms: Utilizing the latest geometric deep learning techniques, GeoEnzyme can accurately and efficiently select the most promising starting enzymes.
-
Multi-Objective Screening: Capable of simultaneously evaluating various enzyme characteristics, ensuring the identification of ideal enzymes that meet multiple requirements for complex application scenarios.
-
Built-in Enzyme Database: Provides a well-organized built-in enzyme database managed by EC numbers, facilitating a smooth enzyme mining process.
Inputs¶
To submit an Enzyme Mining job, please open the Project Editor and click "New Job" button on the left sidebar. Then click "Enzyme Mining" under the "Protein Design" group to open the job submission page.

-
Input Mode: The input mode is divided into "Enzyme List" and "EC Number." "Enzyme List" allows users to provide a candidate enzyme library, while "EC Number" enables users to utilize the default candidate enzyme library by specifying an "EC Number."
-
Enzyme List: Click the "
" button to upload a FASTA file as the candidate enzyme library for screening.
-
EC Number: Enter an EC number consisting of four numbers separated by dots (e.g., "1.1.1.1") to use the built-in enzyme library in GeoEnzyme corresponding to that EC number.
-
-
Reactants: The reactants in the enzyme-catalyzed reaction, input as SMILES expressions. Each line should contain one SMILES, supporting chiral molecules. Please refer to Using Ketcher for details.
-
Products: The products in the enzyme-catalyzed reaction, with the same input format as Reactants.
-
By-reactants: When the Objective is set to "Stereoselectivity", this field indicates the by-reactants in the enzyme-catalyzed reaction, formatted identically to Reactants. The by-reactants should be identical to the reactants except for chirality; if the reactants are achiral or chirality does not affect the reaction, the by-reactants should match the reactants completely.
-
By-Products: When the Objective is set to "Stereoselectivity", this field specifies the by-products in the enzyme-catalyzed reaction, formatted identically to Reactants. The only difference between the by-products and the products should be in their chiral configuration. If multiple chiral centers are present in the product, please specify the chiral isomers corresponding to the wild-type enzyme-catalyzed products and the main by-products.
-
Job Name: The name of the job. Please note that the job name must be unique within the project.
Models & Parameters¶
You can use our proprietary GeoEnzyme model to run this job. The parameters of this model are as follows:
-
Objective: You can select one or more screening objectives from "Enzyme Activity" "pH Stability" "Thermostability" "Stereoselectivity" and "Solubility".
-
"Enzyme Activity" refers to the screening of enzyme activity;
-
"pH Stability" and "Thermostability" refer to the screening of pH stability and thermostability of the enzyme, respectively, by specifying the conditions of the enzyme-catalyzed reaction system.
-
"Stereoselectivity" refer to the screening of stereoselectivity, where the chirality of the product and the main by-product must be specified in the input.
-
"Solubility" refers to the screening of enzyme solubility, with the default expression system being Escherichia coli.
-
-
pH: If Objective includes "pH Stability", you can specify the pH value in the reaction system, which can range from 0 to 14, with a default value of 7.
-
Temperature: If Objective includes "Thermostability", you can specify the temperature in the reaction system, which can range from 0 to 100, with a default value of 37.
Results¶
Click Job Results in the Files & Jobs panel to view the job results.
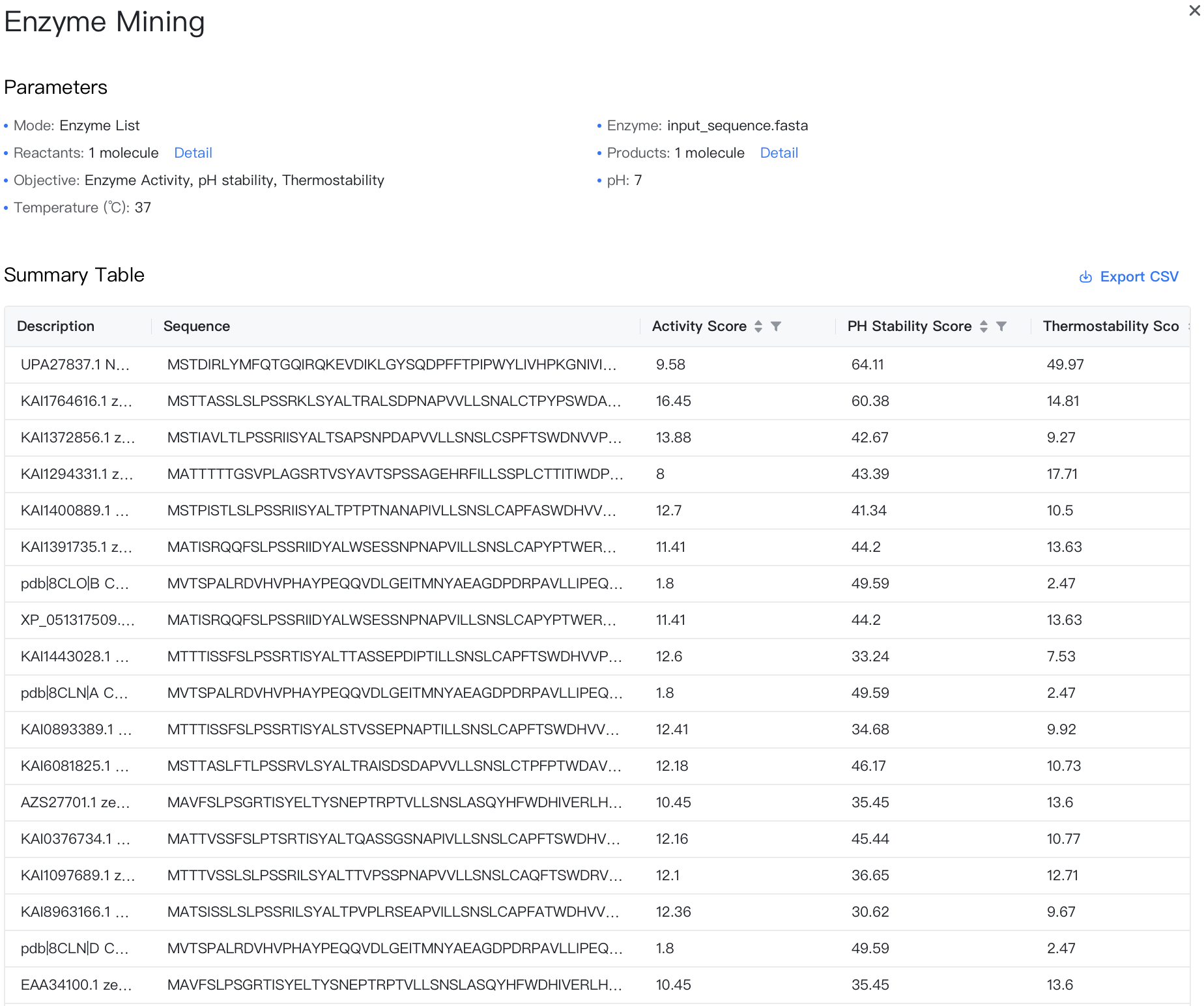
Summary Table¶
The results are stored in a CSV file, which can be downloaded by clicking the "" button to the top-right of the Summary Table.
The summary table contains the following columns:
-
Description: A brief description of the enzyme used to differentiate between different enzymes.
-
Sequence: The enzyme sequences to be screened.
-
Activity Score: A score indicating the enzyme activity.
-
pH Stability Score: A score indicating the stability at a specified pH; higher values indicate better stability.
-
Thermostability Score: A score indicating the stability at a specified temperature; higher values indicate better stability.
-
Stereoselectivity Score: A score indicating the stereoselectivity; higher values indicate a greater proportion of the target chiral product in the total product.
-
Ecoli Solubility Score: A score indicating the solubility in Escherichia coli; higher values indicate better solubility.