Enzyme Optimization¶
Enzyme optimization is the process of optimizing the enzymatic properties of an enzyme, including Enzyme Activity, pH Stability, Thermostability, Stereoselectivity and Solubility, based on its sequence. By analyzing the impact of single or multiple point mutations of enzymes on their enzymatic properties, users can be provided with targeted evolutionary strategies to achieve enzyme modification for specific functions.
Challenges & Features¶
The catalytic process of enzymes is quite complex, with significant differences in the catalytic mechanisms of different enzymes. They exhibit high selectivity for different substrates and different stereoisomers of the same substrate. Additionally, the catalytic ability of enzymes is greatly influenced by environmental conditions such as temperature and pH, making it difficult for traditional methods to consider all these factors simultaneously. In this regard, the enzyme optimization module in GeoBiologics offers the following advantages:
-
Advanced Algorithms: GeoEnzyme utilizes the latest geometric deep learning techniques to analyze the interactions between the entire enzymatic reaction and the enzyme itself. In particular, GeoEnzyme takes into account the chiral selectivity of enzymes toward small molecular reactants and products, providing more accurate predictions.
-
Support for Reaction Conditions: GeoEnzyme considers the impact of different temperatures and pH levels on the enzymatic process. Users can specify reaction conditions such as temperature and pH, allowing for simulations that better reflect real-world applications.
-
User-Friendly: GeoEnzyme supports the simultaneous screening of specified single-point and multi-point mutations. You can clearly compare the effects of different mutation strategies on enzymatic properties and iteratively refine the direction of enzyme evolution.
Inputs¶
To submit an Enzyme Optimization job, please open the Project Editor and click "New Job" button on the left sidebar. Then click "Enzyme Optimization" under the "Protein Design" group to open the job submission page.

-
Enzyme Sequence: The enzyme sequence to be optimized, with a maximum length of 1000 amino acids.
-
Reactants: The reactants in the enzyme-catalyzed reaction, input as SMILES expressions. Enter one SMILES expression per line, supporting chiral molecules. Please refer to Using Ketcher for details.
-
Products: The products in the enzyme-catalyzed reaction, with the same input format as Reactants.
-
By-reactants: When the Objective is set to "Stereoselectivity", this field indicates the by-reactants in the enzyme-catalyzed reaction, formatted identically to Reactants. The by-reactants should be identical to the reactants except for chirality; if the reactants are achiral or chirality does not affect the reaction, the by-reactants should match the reactants completely.
-
By-Products: When the Objective is set to "Stereoselectivity", this field specifies the by-products in the enzyme-catalyzed reaction, formatted identically to Reactants. The only difference between the by-products and the products should be in their chiral configuration. If multiple chiral centers are present in the product, please specify the chiral isomers corresponding to the wild-type enzyme-catalyzed products and the main by-products.
-
Mut. mode: The mutation mode can be either "Saturation Mutagenesis" or "Custom". "Saturation Mutagenesis" indicates single-site saturation mutations, while "Custom" allows for custom single or multiple combinations of mutations.
-
Mutation sites: When the mutation mode is "Saturation Mutagenesis", mutation sites must be entered. Use commas to separate entries, e.g., 32-36 or 100,102-105, where 32-36 represents a closed interval, including sites 32, 33, 34, 35, and 36.
-
Mutation list: When the mutation mode is "Custom", a mutation list must be provided. Each line represents a single or multiple mutations, with multiple mutations separated by commas, e.g., G25T (single mutation) or A35W,Q64F,E103T (three mutations).
-
-
Job Name: The name of the job. Please note that the job name must be unique within the project.
Models & Parameters¶
You can use our proprietary GeoEnzyme model or a custom-trained model from the Enzyme Optimization (Training) (where the model name matches the training task name) to run this job. The parameters of the model are as follows:
-
Objective: You can select one or more optimization objectives from "Enzyme Activity" "pH Stability" "Thermostability" and "Stereoselectivity"
-
"Enzyme Activity: refers to the optimization of enzyme activity;
-
"pH Stability" and "Thermostability" refer to the optimization of pH stability and thermostability of the enzyme, respectively, by specifying the conditions of the enzyme-catalyzed reaction system.
-
"Stereoselectivity" refer to the optimization of stereoselectivity, where the chirality of the product and the main by-product must be specified in the input.
-
"Solubility" refers to the optimization of enzyme solubility, with the default expression system being Escherichia coli.
-
-
pH: If Objective includes "pH Stability", you can specify the pH value in the target system, which can range from 0 to 14, with a default value of 7.
-
Temperature: If Objective includes "Thermostability", you can specify the temperature in the target system, which can range from 0 to 100, with a default value of 37.
Results¶
Click Job Results in the Files & Jobs panel to view the job results.
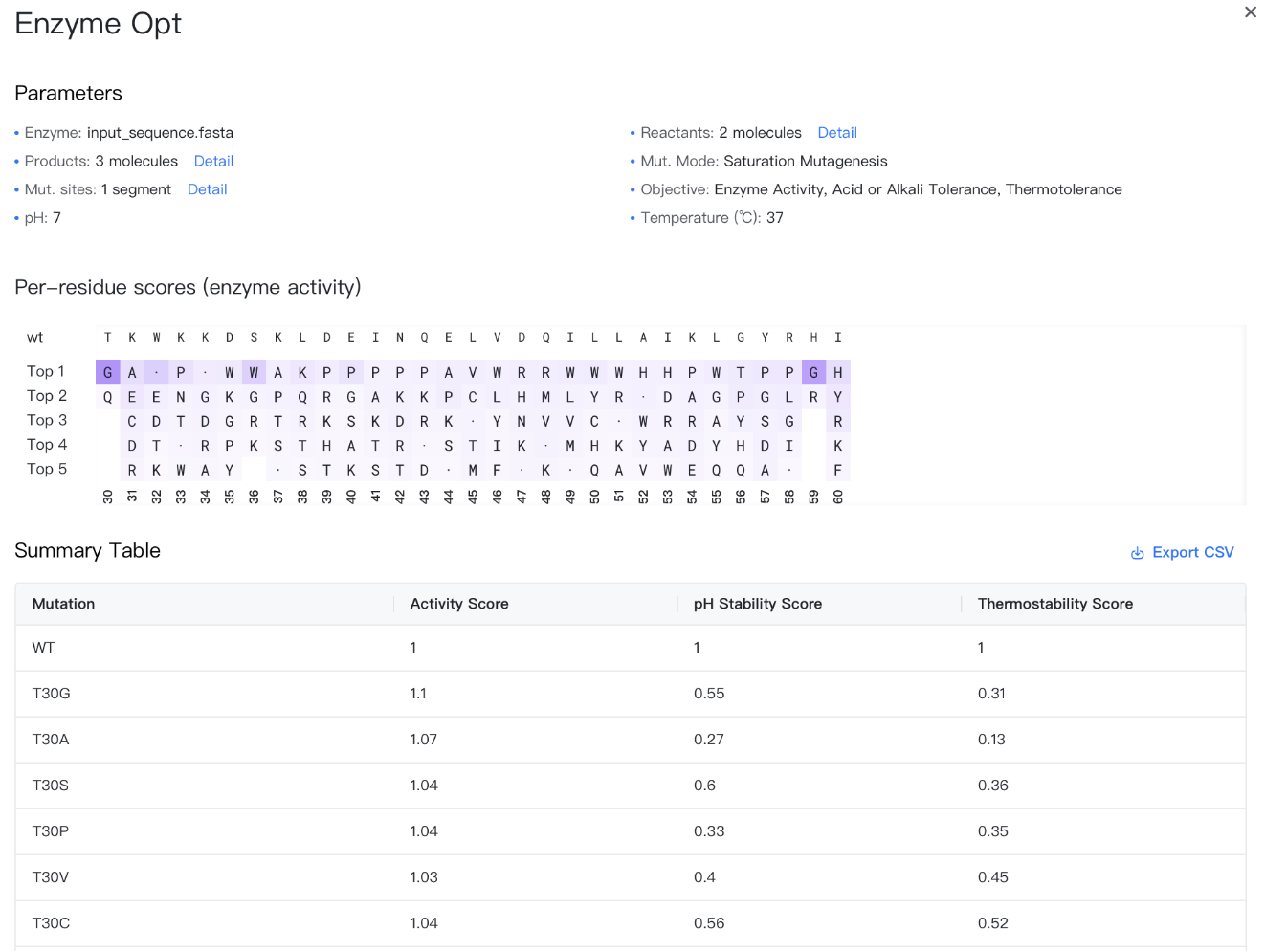
Summary Table¶
The results are stored in a CSV file, which can be downloaded by clicking the "" button to the top-right of the Summary Table.
The summary table contains the following columns:
-
Mutation: Labels of the point mutations, formatted as "original amino acid + residue ID + mutated amino acid", e.g., F103Y. The first row is fixed as "WT", representing the wild-type enzyme before mutation.
-
Activity Score: Changes in enzyme activity caused by mutations; a score greater than 0 indicates increased activity, less than 0 indicates decreased activity, and equal to 0 indicates no change. The score for the first row "WT" is fixed at 0.
-
pH Stability Score: Changes in the pH stability due to mutations, representing enzyme activity at the specified pH; a score greater than 0 indicates improved pH stability, less than 0 indicates decreased stability. The score for the first row "WT" is fixed at 0.
-
Thermostability Score: Changes in the thermostability due to mutations, representing enzyme activity at the specified temperature; a score greater than 0 indicates improved thermostability, less than 0 indicates decreased thermostability. The score for the first row "WT" is fixed at 0.
-
Ecoli Solubility Score: Changes in the solubility due to mutations; a score greater than 0 indicates improved thermal stability, less than 0 indicates decreased stability. The score for the first row "WT" is fixed at 0.
Mutation Suggestions¶
If the mutation mode selected is "saturation mutagenesis", the results page will also include a section "Per-residue scores" according to "Objective", shows the top 5 amino acids recommended for each mutation site.
- The "wt" row shows the wild-type amino acid for each mutation site.
- "Top 1-5" lists the top 5 recommended amino acids for each site.
- Mutant amino acids with lower scores are not displayed, except for self-mutations.
- Mutant amino acids are displayed in each cell. Self-mutations are marked with "·" for easier identification.
- The background color of each cell shows the affinity advantage of the current amino acid at the current mutation site among all 20 possible amino acids. If the background color of the self-mutation is light, it indicates that the site has a large potential for enzyme modification.
When you hover over a cell, a tooltip will appear showing the wild-type and mutant amino acids.